Articles
- Page Path
- HOME > Sci Ed > Volume 1(1); 2014 > Article
-
Original Article
An ontology-based biomedical research paper authoring support tool -
Senator Jeong1
 , Sejin Nam2
, Sejin Nam2 , Hyun-Young Park1
, Hyun-Young Park1
-
Science Editing 2014;1(1):37-42.
DOI: https://doi.org/10.6087/kcse.2014.1.37
Published online: February 13, 2014
1National Center for Medical Information and Knowledge, Korea National Institute of Health, Cheongwon, Korea
2Biomedical Knowledge Engineering Laboratory, Seoul National University, Seoul, Korea
- Correspondence to Senator Jeong E-mail: senatorjeong@gmail.com
- This paper was posted on the Journal Article Tag Suite Conference proceedings website available from: http://www.ncbi.nlm.nih.gov/books/NBK159968/
• Received: October 4, 2013 • Accepted: December 1, 2013
Copyright © Korean Council of Science Editors
This is an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
- This work aims to develop a paper authoring support system that helps biomedical scientists to organize their ideas for a specific discourse purpose. As an initial step toward the goal, this study developed an abstract authoring support tool that provides candidate lexical bundles organized according to the introduction, methods, results, and discussion (IMRAD) structure. Lexical bundles were extracted from the sentences in 152,083 structured abstracts of the PubMed Central open access subset and their distribution was analyzed by IMRAD section. To organize lexical bundles according to IMRAD, the Lexical Bundle Ontology was built. A Journal Article Tag Suite compliant authoring support tool was implemented. This tool lists candidate lexical bundles corresponding to authors’ discourse purposes in a specific section and thereby helps to complete sentences. We expect this tool be useful, at least in biomedical abstract writing, to organize an author’s ideas to achieve a specific discourse purpose. This tool is targeted primarily at biomedical scientists whose mother tongue is not English; however, English native speakers may find it useful as well.
- Biomedical research papers are typically formatted according to the introduction, methods, results, and discussion (IMRAD) structure. Even with the guidance of this format, writing a paper in English is challenging for authors because the most difficult thing is to organize and present their ideas with appropriate expressions. This is true to some extent for all authors, regardless of whether their mother tongue is English or not. Our goal is to develop a paper authoring support system that helps biomedical scientists to organize their ideas for a specific discourse purpose. As an initial step toward the goal, this study developed an abstract authoring support tool that helps to write sentences using lexical bundles organized according to the IMRAD structure. Lexical bundles function as the basic building blocks of this discourse structure. For example, the lexical bundle “the purpose of this study was” indicates the research purpose in the introduction section.
- The motivation for this study arises from the fact that each IMRAD section has a highly codified discourse purpose and frequently occurring lexical bundles. Our approach also draws on the literature that has determined that (1) communicative meanings and functions are often realized by formulaic expression [1]; and that (2) in complying with IMRAD format, academic authors adopt predetermined phraseological patterns in discourse steps [2].
- One study showed that a more compact set of vocabulary is used in biomedical science texts than in general English [3]. Along the same lines, Wang et al. [4] compiled the medical academic word list, which is a list of the most frequently used medical academic words in medical research papers. However, individual word forms are not sufficient for building an author support tool because most rhetorical functions are not accomplished by choosing a word from a word list.
- Lexical bundles are combinations of three or more words that frequently occur in a corpus [5]. They have many names: ‘n-grams’, ‘multi-word patterns’, ‘formulaic patterns’, ‘clusters’, or ‘collocations’. Though they are not complete structural units, lexical bundles work as basic building blocks of discourse. Several researchers focused on characterizing lexical bundles in research articles. Cortes [6] analyzed the relationship of the lexical bundles to the ‘moves’ in the Introduction section of research articles. He identified that some lexical bundles composed of more than five words trigger the communicative function of a move (e.g., ‘the purpose of this study was’ describes the objective of study). Saber analyzed the distribution of lexical bundles among the different IMRAD sections in biomedical research articles and found a limited repertoire of key standardized phraseological patterns specific to certain rhetorical steps [2]. Daniel [7] demonstrated that lexical bundles work as clear signals of discourse purpose in abstract texts. Lorenzo [8] investigated the frequency, structure, and functions of lexical bundles in English research papers.
- Previous studies have demonstrated the utility of lexical bundles in achieving a specific discourse purpose. A formulaic expression compendium is another example of their utility [9]. Drawing upon the utility of lexical bundles, we developed an abstract writing support tool that can help an author to combine the appropriate lexical bundles with a given discourse step.
Introduction
- Data source
- To collect lexical bundles for the authoring support tool, we identified the salient lexical bundles in a specific IMRAD section. The data source for the lexical bundles was the structured abstracts of research articles (n=152,083) in the PubMed Central open access subset [10]. We repurposed the data collected for our another project which classify sentences in biomedical paper abstracts. Therefore, the data processing steps in this study are partially the same as those in that project.
- Extraction of lexical bundles
- Our structured abstract corpus contains a variety of section headings (n=1,628). These variants include plurals (conclusion, conclusions), modifiers (conclusion, major conclusion), different word sequences (conclusions and significance, significance and conclusion), and joint section headings (method and result, result and discussion) [11]. Different section headings representing the same concept were merged into 1,001 headings using OpenRefine (http://openrefine.org), then further normalized into IMRAD using the National Library of Medicine’s category mapping table [12]. The mapping table uses five categories: background, objective, methods, results, and conclusions. In our study, background and objective were merged into introduction and conclusions was renamed to discussion. To extract lexical bundles, LingPipe was used for sentence splitting [13]. Each sentence was appended with its corresponding IMRAD heading.
- After grouping the sections into IMRAD, lexical bundles were extracted from the sentences in each section, and then their occurrence frequency was analyzed using our in-house program. The lexical bundles with an occurrence frequency of under 50 instances were ignored. The lexical bundles in this study are defined as: all n-grams (from 2- to 10-gram) without any further processing except numbers, which were marked with a special tag, NUM. Table 1 shows that each IMRAD section has commonly occurring lexical bundles.
- Lexical bundle ontology
- We designed a simple ontology, Lexical Bundle Ontology (LBO), that organizes lexical bundles according to IMRAD. Thus, LBO extends the generic IMRAD model to produce its own classes. LBO has four classes under the top class Lexical- Bundle: IntroductionBundle, MethodBundle, ResultBundle, and DiscussionBundle. Each class has three datatype properties: frequency, nGram, and rdfs:label. The prefix ‘lbo’ was declared for the dereferenceable uniform resource identifier (http://lbo.studiosusan.kr/), where an application can find and access bundle instances. The box below shows an LBO instance in the Turtle format. The 9-gram lexical bundle (‘The aim of the present study was to evaluate’) is identified by ‘IntroLBl1108’ and its frequency is 326.
- Implementation of authoring support tool
- A Journal Article Tag Suite (JATS)-compliant authoring support tool prototype was implemented. This proof-of-concept tool was designed to list candidate lexical bundles corresponding to a writer’s discourse purpose and thereby help to complete sentences in the abstract. The Apache Lucene library was used for indexing and searching the lexical bundles in the LBO. To obtain recommend lexical bundles for a specific discourse purpose, they can be sorted by occurrence frequency, length of n-gram, or alphabetical code order. In our tool, the bundles are sorted by their length or occurrence frequency.
- A user completes a sentence by juxtaposing the last words of a given bundle with the first words of other bundles available to come. To make it happen, the left token-first-partial matching method was applied. When a user types a string, the tool matches bundles from the index file in real-time and returns the matched items to the user. For an interactive user interface, AJAX was used.
Methods
Section grouping and sentence splitting
Analysis of lexical bundles
:IntroLbl1108 a :IntroductionBundle; rdfs:label “The aim of the present study was to evaluate”; :frequency 326; :nGram 9.
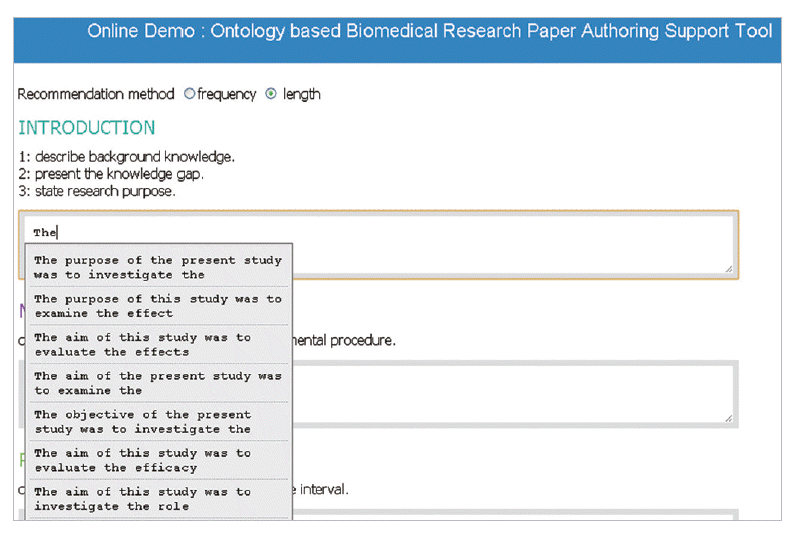
- When a user writes an abstract according to the IMRAD format, the tool lists up to 10 candidate lexical bundles (Fig. 1). The tool is case-sensitive. When a user enters an uppercase letter at the very beginning of a string, the tool returns all bundles beginning with the typed letter. The completed texts are transformed to JATS format by clicking the button labeled ‘Convert to JATS format.’ The Paper Authoring Support Tool is available at http://147.46.70.42:7000/jats/demo.html.
Results
- In this section, we present a use-case scenario to show the utility of the tool for authors. Let us demonstrate the writing of the abstract of a published paper [14] with this tool.
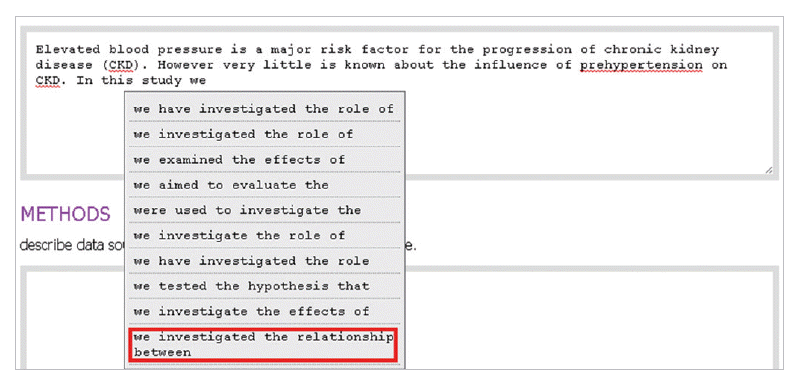
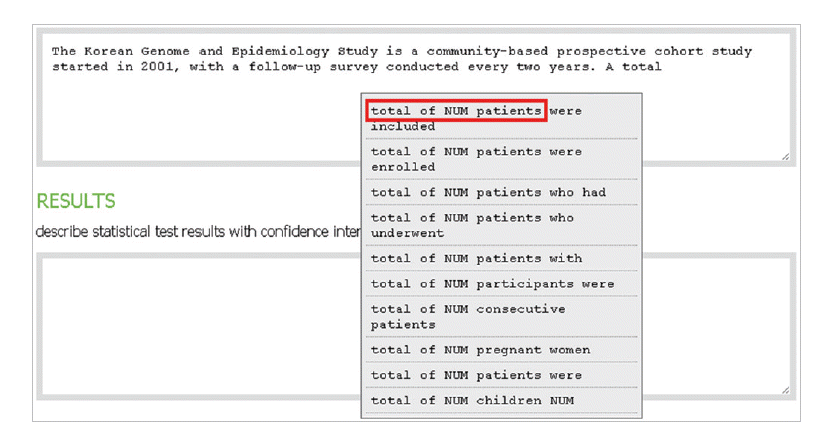
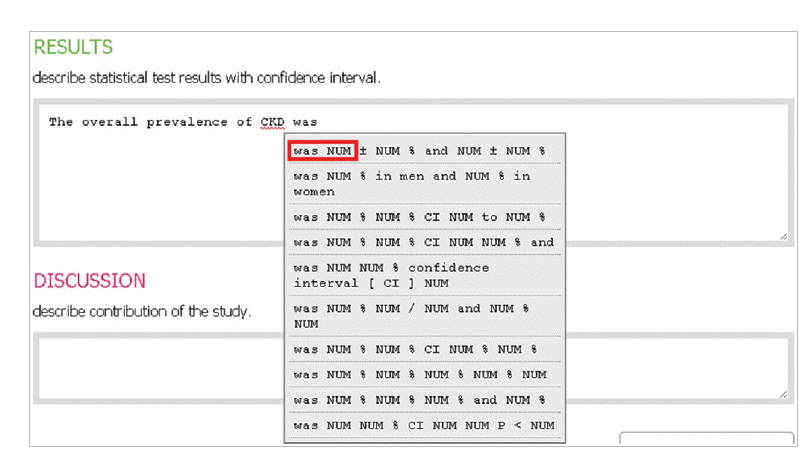
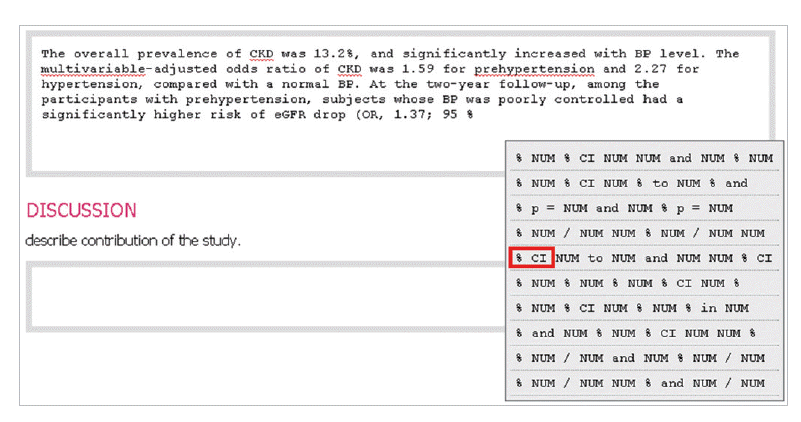
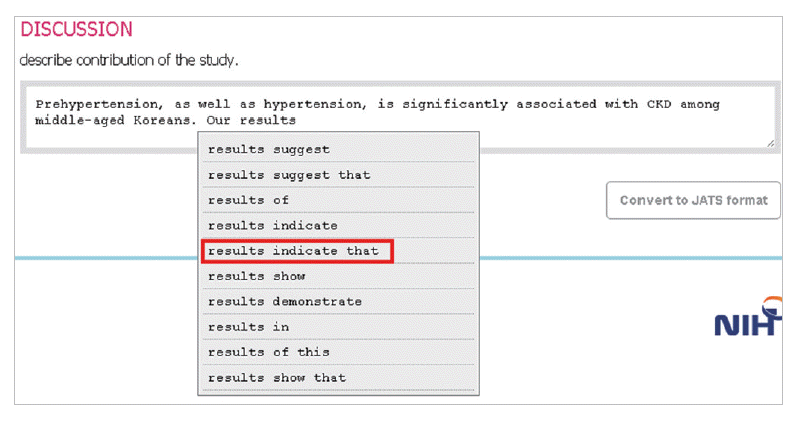
- In the introduction section, the author begins by describing background knowledge and presenting the knowledge gap with the lexical bundle ‘However little is known about’ (Fig. 2). Then she states the research purpose with an expression such as ‘In this study we investigated the’ (Fig. 3). The methods section often involves describing a study type (Fig. 4), subject (Fig. 5), and experimental procedure (Fig. 6). The results section often has statistical test results (Fig. 7) with a confidence interval (Fig. 8). The discussion section often ends with an expression such as ‘Our results indicate that’ (Fig. 9). In each step of writing, the author selects a proper bundle serving her discourse purpose. Finally, by clicking the appropriate button, the author can transform the text to the JATS extensible mark-up language.
Use-case scenario
- This study presented an abstract authoring support tool that lists candidate lexical bundles responding to authors’ discourse purposes in a specific IMRAD section and can help to complete sentences. We hope the use case scenario has demonstrated the viability of this proof-of-concept tool.
- The implications of the tool are obvious. It would be a useful tool, at least in abstract writing, to organize the author’s ideas to achieve a specific discourse purpose. It would thereby generate a well-organized and clearly written manuscript. This tool is targeted primarily at biomedical scientists whose mother tongue is not English. However, native speakers may still find it useful. This tool could also be useful for authors in any other scientific domain because many lexical bundles are commonly used in papers across the sciences.
- Admittedly, this exploratory study extracted the lexical bundles from the abstracts in the PubMed Central open access subset; thus it does not show all of the thematic contents in the bodies of research papers. Further studies are required to cover full-text papers. Further work would include refining the authoring support tool. Grouping the lexical bundles with a similar role into a single group would be one approach (e.g., the two lexical bundles ‘the objective of this study’ and ‘the aim of this study’ have a similar role). This semantic structure will assist in highly selective searching for lexical bundles.
- In conclusion, we hope that the present work will provoke the science community to start discussions on and endorsement of this authoring support tool.
Discussion
-
Acknowledgements
- This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (no. 2011-0010515) and by the Korea National Institute of Health (no. 4800-4848-307 and no. 3000-3334-300).
Fig. 2.The author begins by describing background knowledge and presenting the knowledge gap with the lexical bundle, ‘However little is known about.’
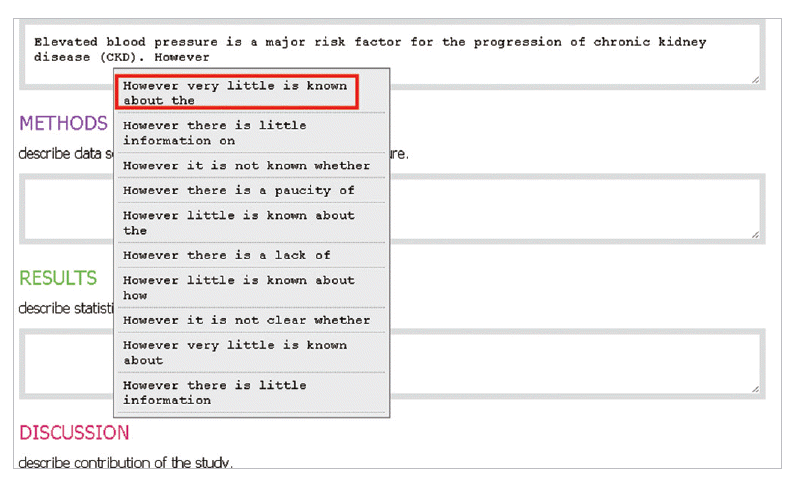

Fig. 3.The author states research purpose with a lexical bundle, ‘In this study we investigated the.’
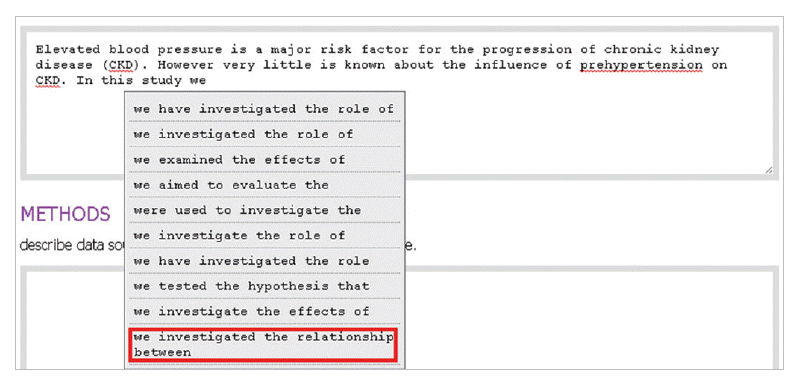

Fig. 4.The author describes a study type by selecting a lexical bundle, ‘prospective cohort study’ matching with her discourse purpose.
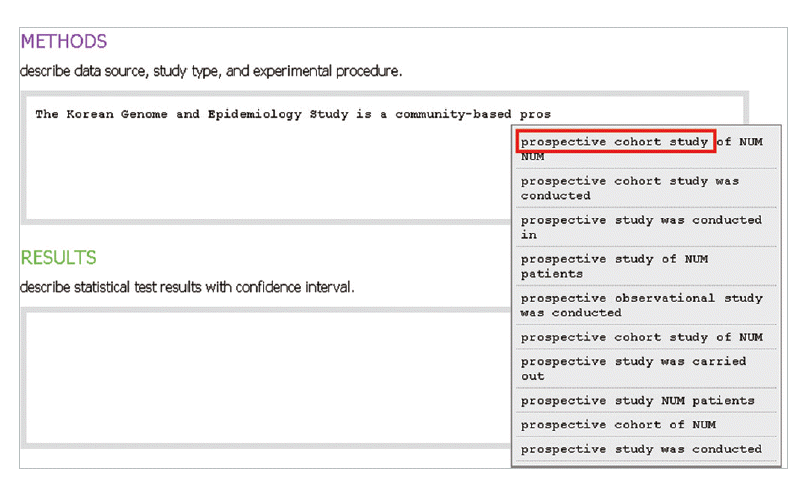

Fig. 5.In the methods section, the author describes experimental subject with a lexical bundle, “total of NUM patients.”
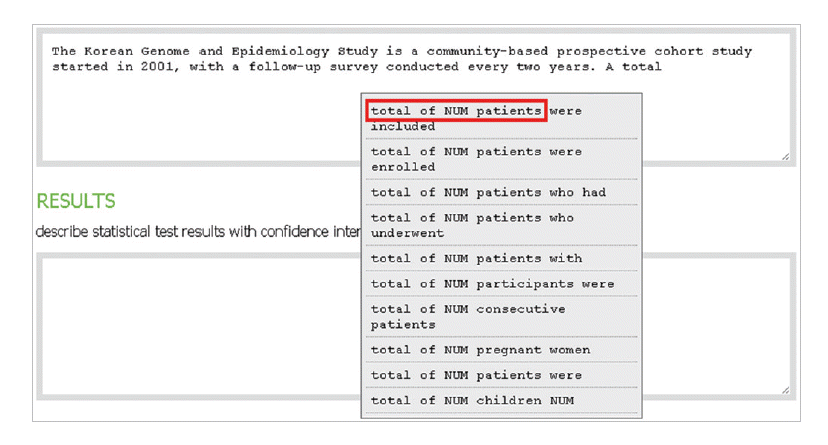

Fig. 6.In the methods section, the author describes experimental procedure with a lexical bundle, ‘classified according to the.’


Fig. 9.The author describes the discussion section with an lexical bundle, ‘Our results indicate that.’
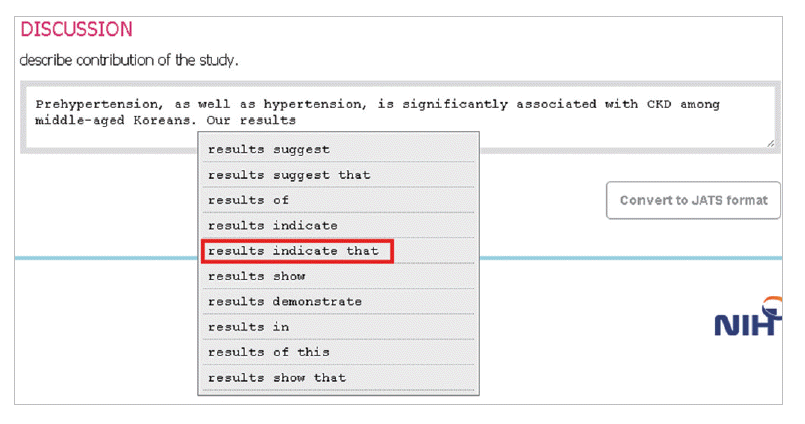

Table 1.Distribution of the top 3 lexical bundles in each IMRAD (introduction, methods, results, and discussion) section of the PubMed Central open access subset (from 2- to 10-gram bundles)
- 1. Martinez R, Schmitt N. A phrasal expressions list. Appl Linguist 2012;33:299-320.http://dx.doi.org/10.1093/applin/ams010. ArticlePDF
- 2. Saber A. Phraseological patterns in a large corpus of biomedical articles. In : Boulton A, Carter-Thomas S, Rowley-Jolivet E, editors. Corpus-informed research and learning in ESP: issues and applications. Amsterdam: John Benjamins Publishing Company; 2012:45-82.Article
- 3. Waxmonsky S, Goldsmith J, Rzhetsky A. Discovering and counting biomedical verbs. Paper presented at: 2010 Ninth International Conference on Machine Learning and Applications (ICMLA). 2010 Dec 12-14; Washington, DC, USA. Article
- 4. Wang J, Liang SI, Ge GC. Establishment of a medical academic word list. Engl Specif Purp 2008;27:442-58.http://dx.doi.org/10.1016/j.esp.2008.05.003. Article
- 5. Biber D, Conrad S, Cortes V. If you look at …: lexical bundles in university teaching and extbooks. Appl Linguist 2004;25:371-405.http://dx.doi.org/10.1093/applin/23.371. ArticlePDF
- 6. Cortes V. The purpose of this study is to: connecting lexical bundles and moves in research article introductions. J Engl Acad Purp 2013;12:33-43.http://dx.doi.org/10.1016/j.jeap.2012.11.002. Article
- 7. Daniel R. Domain-independent mining of abstracts using indicator phrases. DLib Mag 2012;18:http://dx.doi.org/10.1045/july2012-daniel. Article
- 8. Lorenzo SD. Lexical bundles in scientific English: a corpus-based study of native and non-native writing [dissertation]. Barcelona: Universitat de Barcelona; 2011.
- 9. Morley J. Academic Phrasebank [Internet]. [place unknown]: University of Manchester; 2013 [cited 2013 Apr 6]. Available from: http://www.phrasebank.manchester.ac.uk.
- 10. US National Library of Medicine. PMC: FTP service [Internet]; Bethesda: US National Library of Medicine; [cited 2011 Apr 12]. Available from: http://www.ncbi.nlm.nih.gov/pmc/tools/ftp/.
- 11. Ripple AM, Mork JG, Rozier JM, Knecht LS. Structured abstracts in MEDLINE: twenty-five years later [Internet]. Bethesda: US National Library of Medicine; 2012 [cited 2013 Dec 29]. Available from: http://structuredabstracts.nlm.nih.gov/Structured_Abstracts_in_MEDLINE_Twenty-Years_Later.pdf.
- 12. USA.gov. Structured abstracts in MEDLINE: implementation information [Internet]; Bethesda: US National Library of Medicine; 2012 [cited 2012 Sep 22]. Available from: http://structuredabstracts.nlm.nih.gov/Implementation.shtml.
- 13. Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics 2004;20:2320-1.http://dx.doi.org/10.1093/bioinformatics/bth227. ArticlePDF
- 14. Kim MJ, Lim NK, Park HY. Relationship between prehypertension and chronic kidney disease in middle-aged people in Korea: the Korean genome and epidemiology study. BMC Public Health 2012;12:960. http://dx.doi.org/10.1186/1471-2458-12-960. ArticlePubMedPMCPDF
References
Figure & Data
References
Citations
Citations to this article as recorded by 

- Structuralizing biomedical abstracts with discriminative linguistic features
Sejin Nam, Senator Jeong, Sang-Kyun Kim, Hong-Gee Kim, Victoria Ngo, Nansu Zong
Computers in Biology and Medicine.2016; 79: 276. CrossRef - Editing and publishing scholarly journals in the internet age
Kihong Kim
Science Editing.2014; 1(1): 2. CrossRef

 KCSE
KCSE


 PubReader
PubReader ePub Link
ePub Link Cite
Cite